MethGo
DNA methylation is a major epigenetic modification regulating several biological processes. A standard approach in the study of DNA methylation is bisulfite sequencing (BS-Seq). BS-Seq couples bisulfite conversion of DNA with next generation sequencing to provide a genome wide profile of DNA methylation at single base resolution. The analysis of BS-Seq data involves the use of customized aligners for mapping reads and additional bioinformatic pipelines for downstream data analysis. While most post-alignment programs generate methylation calls, MethGo carries out subsequent genomic and epigenomic analyses to comprehensively explore BS-Seq datasets.
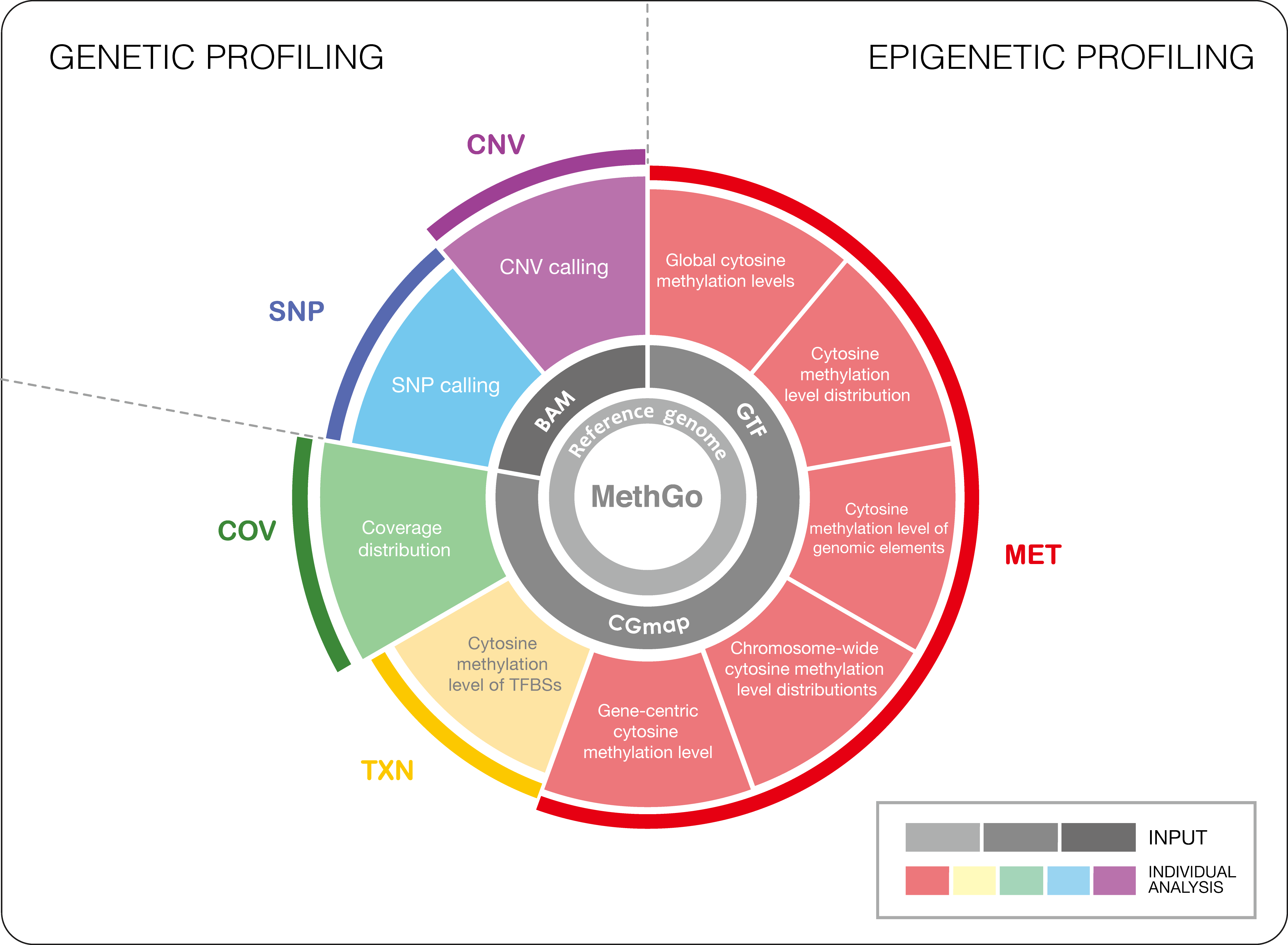
MethGo is a simple and effective tool designed for the analysis of data from whole genome bisulfite sequencing (WGBS) and reduced representation bisulfite sequencing (RRBS). MethGo provides 5 major modules:
- COV: Coverage distribution of each cytosine
- MET: Both global and gene-centric cytosince methylation levels
- TXN: Cytosine methylation levels at transcription factor binding sites (TFBSs)
- SNP: Single nucleotide polymorphism (SNP) calling
- CNV: Copy number variation calling
MethGo is available under the terms of the MIT license.